Navigation
Illuminating Study Reveals How Plants Respond to Light
Most of us take it for granted that plants respond to light by growing, flowering and straining towards the light, and we never wonder just how plants manage to do so. But the ordinary, everyday responses of plants to light are deceptively complex, and much about them has long stumped scientists.
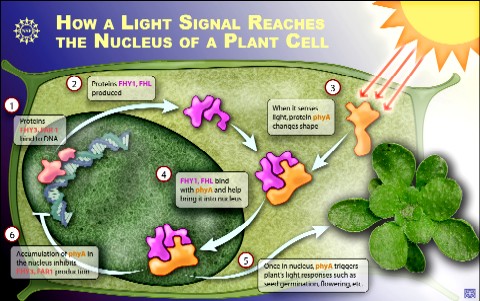 |
| Plants prepare to respond to light while still in the dark. Two proteins known as FHY3 and FAR1 bind to DNA in a plant cell and cause production of two other proteins known as FHY1 and FHL. When hit with far red light, a light-sensitive protein called phytochrome A (phyA) changes its shape. This shape change allows it to bind to FHY1 and FHL. FHY1 and FHL then carry the activated phyA into the cell nucleus. From there, phyA is able to initiate the plant's developmental responses to light such as growth, flowering and straining towards the light. Credit: Zina Deretsky, National Science Foundation |
Most of us take it for granted that plants respond to light by growing, flowering and straining towards the light, and we never wonder just how plants manage to do so. But the ordinary, everyday responses of plants to light are deceptively complex, and much about them has long stumped scientists.
Now, a new study "has significantly advanced our understanding of how plant responses to light are regulated, and perhaps even how such responses evolved," says Michael Mishkind, a program director at the National Science Foundation (NSF) which funded this study. The study is published in the November 23, 2007 issue of Science.
Higher plants “learn” about their surroundings from the color, intensity, direction, and duration of light around them, which determine when seeds will sprout; how fast, how tall and in which direction seedlings will grow; and when plants will flower—all traits with practical implications for agriculture. Despite the importance of these processes, scientists are only just beginning to understand how plants perceive and respond to light.
Researchers at the Boyce Thompson Institute for Plant Research on the campus of Cornell University, headed by Assistant Scientist Haiyang Wang, an adjunct assistant professor in Plant Biology at Cornell, in this study announce significant progress towards understanding a cascade of signal-transduction events involving far-red light perception in higher plants.
Light is absorbed by photoreceptors, causing their activation. The photo-activated receptors pass messages to other molecules, which relay them to others, and so on. The effects of the light signal are eventually manifested in the nucleus as changes in activity of DNA-binding proteins, i.e. transcription factors that specifically interact with the regulatory regions of genes, modulating their expression and leading to changes in plant morphology. How the light signal is transduced into the nucleus was poorly understood, despite intensive research efforts.
By conducting experiments with Arabidopsis--a small flowering plant widely used as a model organism--the researchers discovered that the plant prepares to respond to light while it is still in the dark, even before it is exposed to light.
This preparation involves producing a pair of closely related proteins known as FHY1 and FHL in darkness. The researchers found that another pair of closely related proteins named FHY3 and FAR1 act as transcription factors to promote the production of FHY1 and FHL in darkness.
 |
| This image shows micrographs of plant roots. The green dots inside indicate presence of the FHY3 protein in the cells' nuclei. Notice the protein has accumulated in the division zone of the primary root. The artists have added computational models in the middle showing FHY3 protein (in green) binding to DNA (shown in gray). Researchers have discovered that FHY3 and FAR1, two proteins derived from what are called "jumping genes", put into motion the sequence of events that bring about the light responses of a plant such as growth, flowering and straining towards light. Credit: Pelkie, Daniel Ripoll, and Rongcheng Lin |
When plants are exposed to light, the photoreceptor protein phytochrome A is activated through a change in shape that allows it to bind to FHY1 and FHL. The binding of FHY1 and FHL to phytochrome A helps to bring phytochrome A into the nucleus to regulate the activity of genes located in the cell nucleus that govern plant growth and development.
Haiyang Wang, a member of the research team from Boyce Thompson Institute for Plant Research, says that the plant probably stockpiles these proteins (FHY3, FAR1, FHY1, FHL and phytochrome A) needed for light responses in the dark for the same reason that a traveler fills his car's gas tank the night before a morning journey: in order to be able to get going, without delay, at first light.
Moreover, the researchers also discovered the existence of a negative feedback loop between accumulations of phytochrome A in the cell nucleus and the FHY3 and FAR1 proteins that prime the plant's light response system: the more phytochrome A accumulates in the nucleus, the less FHY3 and FAR1 proteins are produced, and so less phytochrome A is imported into the nucleus. "This feedback loop serves as a built-in brake that limits the flow of light responses," says Wang.
In addition to improving understanding of light-signal transduction in Arabidopsis, “This research provides insights into how light responses are controlled in crop plants as well, which may ultimately enhance our ability to produce crops that better fit their ambient light environment through agro-biotechnology,” noted Rongcheng Lin, the lead author.
Collaborator Cédric Feschotte (University of Texas at Arlington) added, “This study has broad implications for evolutionary biology.” The FHY3 and FAR1 proteins are evolutionarily related to Mutator-like transposases—enzyme products of transposable elements, commonly called “jumping genes”—that mediate transposition or jumping of the DNA elements in the genome.
Discovered by Barbara McClintock, a Nobel Laureate and Cornellian (class of 1927), over half a century ago in corn, transposable elements are now known to be widespread among the genomes of many plants and animals. McClintock viewed them as “controlling elements” that provide crucial control over an organism’s key developmental programs, whereas others have regarded them as merely “selfish” or “junk” DNA. Through lack of direct evidence, the “controlling elements” theory has remained controversial.
“This study provides experimental evidence that transposases produced by ancient Mutator-like transposable elements evolved into transcription factors to regulate light-responsive gene expression in higher plants,” explained Feschotte. Of note, such transposable element-derived genes appear to be restricted to the genomes of flowering plants, now the most advanced plant group on Earth. Transformation of ancient transposases into transcription factors likely happened early in the evolution of flowering plants. Together with the innovation of phyA before the origin of flowering plants, this event may have been a key step in acquiring necessary phyA-signaling intermediates, thus contributing to the successful establishment of flowering plants. “Further research will be needed to test this hypothesis,” said Wang.
Other members of the research team include: Dr. Rongcheng Lin and Dr. Lei Ding, who performed most of the experimental work; Dr. Cédric Feschotte and Dr. Claudio Casola, collaborators from the University of Texas at Arlington, who played an essential role in data mining and the phylogenetic analysis of the relationship between the proteins FHY3 and FAR1 with transposases; and Daniel Ripoll, a research scientist in Cornell's Computational Biology Service Unit and a co-author on the Science paper, who played a key role in building computational models of the proteins.
Authors:
Haiyang Wang, Boyce Thompson Institute for Plant Research
And
Lily Whiteman, National Science Foundation
Program Contacts
Michael Mishkind, National Science Foundation
(1) 703-292-8413 mmishkind@nsf.gov
Haiyang Wang, Boyce Thompson Institute for Plant Research
(1) 607-254-7476 hw75@cornell.edu
Search
Latest articles
Agriculture
- World Water Week: Healthy ecosystems essential to human health: from coronavirus to malnutrition Online session Wednesday 24 August 17:00-18:20
- World Water Week: Healthy ecosystems essential to human health: from coronavirus to malnutrition Online session Wednesday 24 August 17:00-18:20
Air Pollution
- "Water and Sanitation-Related Diseases and the Changing Environment: Challenges, Interventions, and Preventive Measures" Volume 2 Is Now Available
- Global Innovation Exchange Co-Created by Horizon International, USAID, Bill and Melinda Gates Foundation and Others
Biodiversity
- It is time for international mobilization against climate change
- World Water Week: Healthy ecosystems essential to human health: from coronavirus to malnutrition Online session Wednesday 24 August 17:00-18:20
Desertification
- World Water Week: Healthy ecosystems essential to human health: from coronavirus to malnutrition Online session Wednesday 24 August 17:00-18:20
- UN Food Systems Summit Receives Over 1,200 Ideas to Help Meet Sustainable Development Goals
Endangered Species
- Mangrove Action Project Collaborates to Restore and Preserve Mangrove Ecosystems
- Coral Research in Palau offers a “Glimmer of Hope”
Energy
- Global Innovation Exchange Co-Created by Horizon International, USAID, Bill and Melinda Gates Foundation and Others
- Wildlife Preservation in Southeast Nova Scotia
Exhibits
- Global Innovation Exchange Co-Created by Horizon International, USAID, Bill and Melinda Gates Foundation and Others
- Coral Reefs
Forests
- NASA Satellites Reveal Major Shifts in Global Freshwater Updated June 2020
- Global Innovation Exchange Co-Created by Horizon International, USAID, Bill and Melinda Gates Foundation and Others
Global Climate Change
- It is time for international mobilization against climate change
- It is time for international mobilization against climate change
Global Health
- World Water Week: Healthy ecosystems essential to human health: from coronavirus to malnutrition Online session Wednesday 24 August 17:00-18:20
- More than 400 schoolgirls, family and teachers rescued from Afghanistan by small coalition
Industry
- "Water and Sanitation-Related Diseases and the Changing Environment: Challenges, Interventions, and Preventive Measures" Volume 2 Is Now Available
- Global Innovation Exchange Co-Created by Horizon International, USAID, Bill and Melinda Gates Foundation and Others
Natural Disaster Relief
- STOP ATTACKS ON HEALTH CARE IN UKRAINE
- Global Innovation Exchange Co-Created by Horizon International, USAID, Bill and Melinda Gates Foundation and Others
News and Special Reports
- World Water Week: Healthy ecosystems essential to human health: from coronavirus to malnutrition Online session Wednesday 24 August 17:00-18:20
- STOP ATTACKS ON HEALTH CARE IN UKRAINE
Oceans, Coral Reefs
- World Water Week: Healthy ecosystems essential to human health: from coronavirus to malnutrition Online session Wednesday 24 August 17:00-18:20
- Mangrove Action Project Collaborates to Restore and Preserve Mangrove Ecosystems
Pollution
- Zakaria Ouedraogo of Burkina Faso Produces Film “Nzoue Fiyen: Water Not Drinkable”
- "Water and Sanitation-Related Diseases and the Changing Environment: Challenges, Interventions, and Preventive Measures" Volume 2 Is Now Available
Population
- "Water and Sanitation-Related Diseases and the Changing Environment: Challenges, Interventions, and Preventive Measures" Volume 2 Is Now Available
- "Water and Sanitation-Related Diseases and the Changing Environment: Challenges, Interventions, and Preventive Measures" Volume 2 Is Now Available
Public Health
- Honouring the visionary behind India’s sanitation revolution
- Honouring the visionary behind India’s sanitation revolution
Rivers
- World Water Week: Healthy ecosystems essential to human health: from coronavirus to malnutrition Online session Wednesday 24 August 17:00-18:20
- Mangrove Action Project Collaborates to Restore and Preserve Mangrove Ecosystems
Sanitation
- Honouring the visionary behind India’s sanitation revolution
- Honouring the visionary behind India’s sanitation revolution
Toxic Chemicals
- "Water and Sanitation-Related Diseases and the Changing Environment: Challenges, Interventions, and Preventive Measures" Volume 2 Is Now Available
- Actions to Prevent Polluted Drinking Water in the United States
Transportation
- "Water and Sanitation-Related Diseases and the Changing Environment: Challenges, Interventions, and Preventive Measures" Volume 2 Is Now Available
- Urbanization Provides Opportunities for Transition to a Green Economy, Says New Report
Waste Management
- Honouring the visionary behind India’s sanitation revolution
- Honouring the visionary behind India’s sanitation revolution
Water
- Honouring the visionary behind India’s sanitation revolution
- Honouring the visionary behind India’s sanitation revolution
Water and Sanitation
- Honouring the visionary behind India’s sanitation revolution
- Honouring the visionary behind India’s sanitation revolution

